Automated Nuclear Pleomorphism Scoring in Breast Cancer¶
Introduction¶
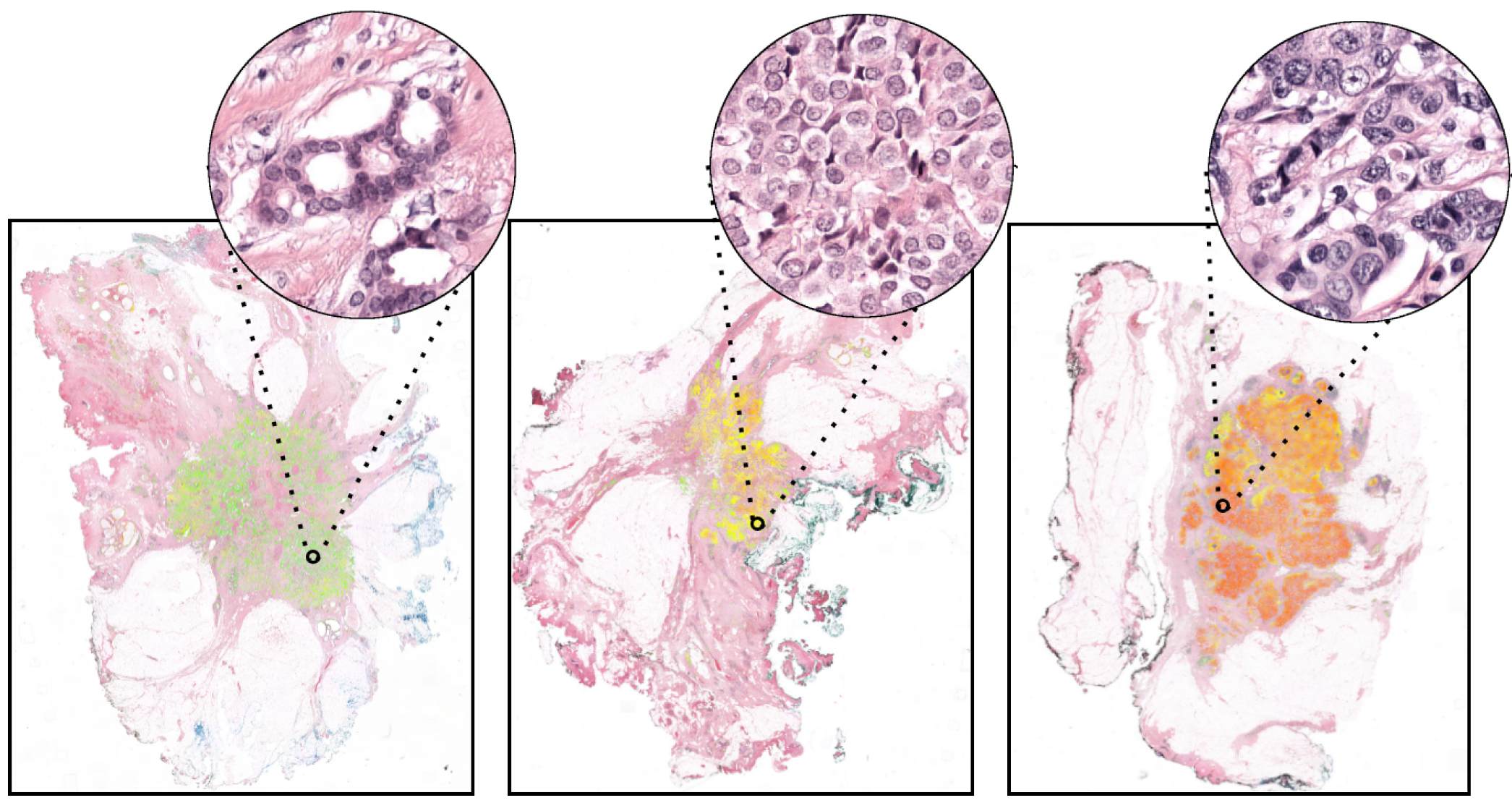
To guide the choice of treatment, every new breast cancer is assessed for aggressiveness (i.e., graded) by an experienced histopathologist. Typically, this tumor grade consists of three components, one of which is the nuclear pleomorphism score (the extent of abnormalities in the overall appearance of tumor nuclei).
The degree of nuclear pleomorphism is subjectively classified from 1 to 3, where a score of 1 most closely resembles epithelial cells of normal breast epithelium and 3 shows the greatest abnormalities. Establishing numerical criteria for grading nuclear pleomorphism is challenging, and inter-observer agreement is poor.
Based on this, in Mercan et al. [1], we studied the use of deep learning to develop fully automated nuclear pleomorphism scoring in breast cancer. The reference standard used for training the algorithm consisted of the collective knowledge of an international panel of 10 pathologists on a curated set of regions of interest covering the entire spectrum of tumor morphology in breast cancer. To fully exploit the information provided by the pathologists, a first-of-its-kind deep regression model was trained to yield a continuous scoring rather than limiting the pleomorphism scoring to the standard three-tiered system. In [1], we showed that our approach achieves top pathologist-level performance in multiple experiments on regions of interest and whole-slide images, compared to a panel of 10 and 4 pathologists, respectively.
Here, we release the test set used in [1], namely the n=118 whole-slide images used in the Slide-study, and implement an automatic evaluation script to allow researchers to submit their predictions on these 118 cases, and benchmark those against the opinion of four pathologists involved in the study, as well as with the reference standard based on the majority vote of their opinion.
Goal¶
The goal of this evaluation platform is to be a reference benchmark for algorithms that can predict nuclear pleomorphism on whole-slide images of breast cancer surgical resections stained in H&E, based on the test data set used in [1]. In line with the idea of challenges in computer vision and medical imaging, here we propose a test data set publicly shared with the scientific community, as well as a benchmarking platform that will allow all researchers to evaluate their algorithm on exactly the same data set and using exactly the same evaluation procedure.
Differently from most challenges in computer vision and medical imaging, we are solely releasing the test set used in [1], together with the evaluation procedure used in that study, implemented on this web platform. The training data used in [1] is not released on this platform and there are currently no plans to release it.
We envision this initiative to be a first step towards promoting research, development, and evaluation of artificial intelligence for the prediction of nuclear pleomorphism in breast cancer and beyond.
How to use this platform¶
To assess the performance of your algorithm on the Slide-study data set used as a test set in [1], you should:
- download the test set from Zenodo at this link; additional details on the dataset can be found on the Data page of this website;
- run your algorithm on the n=118 whole-slide images of this test set
- store the predicted pleomorphism scores in a CSV file following the details of the instructions on the Submission page
- upload your CSV file to this website via a submission; after that, you should be able to see your position on the leaderboard on this website and compare your results with what was reported in [1].
Should you use the data made available via this website and via Zenodo, and the evaluation script implemented on this website, please cite the paper of C. Mercan et al. as follows:
C. Mercan, M. Balkenhol, R. Salgado, M. Sherman, P. Vielh, W. Vreuls, A. Polonia, H. M. Horlings, W. Weichert, J. M. Carter, P. Bult, M. Christgen, C. Denkert, K. van de Vijver, J.-M Bokhorst, J. van der Laak, F. Ciompi, Deep learning for fully-automated nuclear pleomorphism scoring in breast cancer. NPJ Breast Cancer, 2022.
References¶
[1] C. Mercan, M. Balkenhol, R. Salgado, M. Sherman, P. Vielh, W.
Vreuls, A. Polonia, H. M. Horlings, W. Weichert, J. M. Carter, P. Bult,
M. Christgen, C. Denkert, K. van de Vijver, J.-M Bokhorst, J. van der
Laak, F. Ciompi, Deep learning for fully-automated nuclear pleomorphism
scoring in breast cancer. NPJ Breast Cancer, 2022.