Data¶
Data used in this project consists of the Slide-study data set used in [1].
The dataset used in this project case be downloaded from Zenodo.
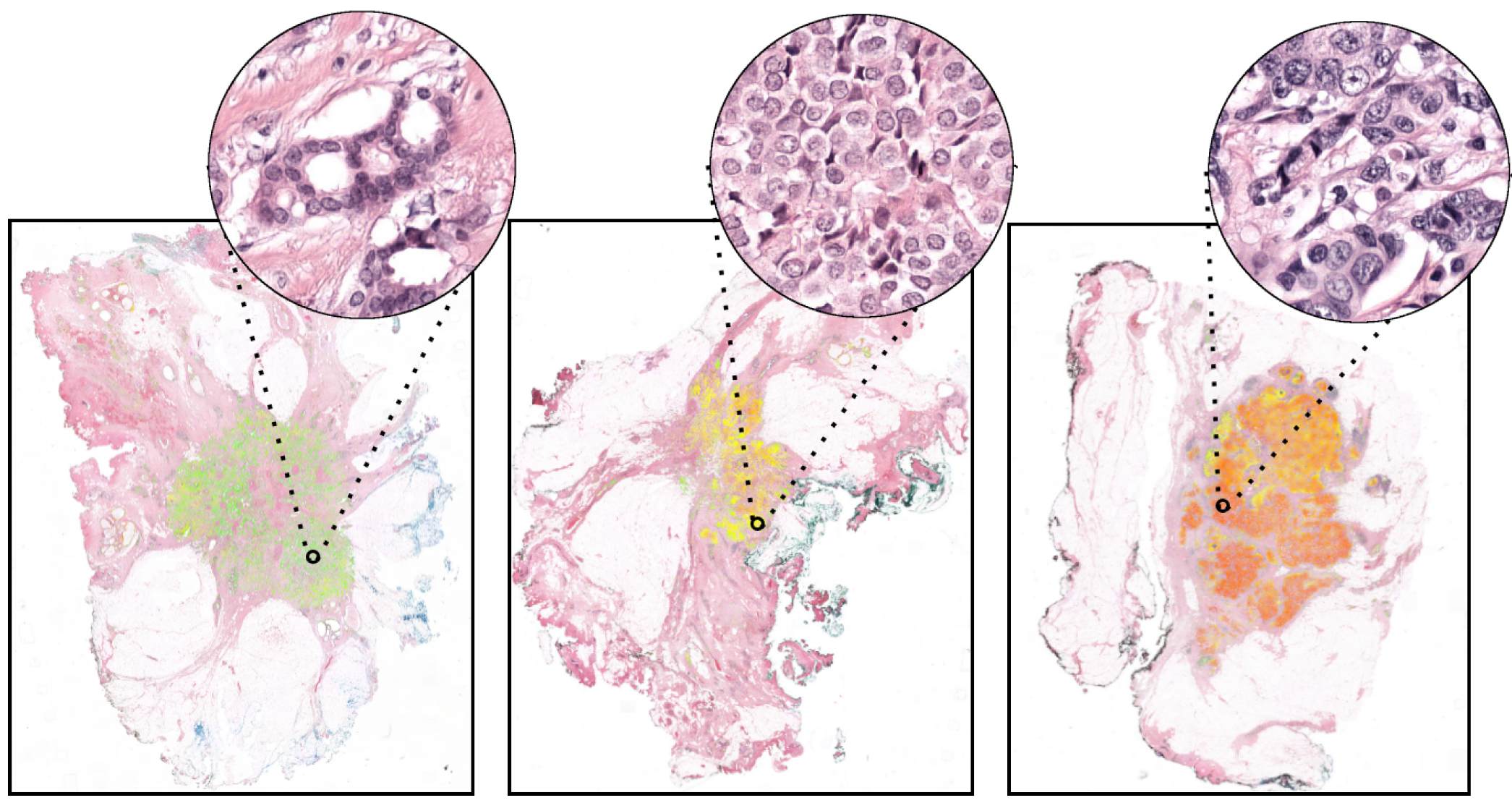
The dataset consists of n=118 digital pathology whole-slide images (WSI) of breast cancer surgical resections, stained with hematoxylin and eosin (H&E) at Radboud University Medical Centers, Nijmegen (The Netherlands).
The WSIs were scanned with a 3DHistech P1000 scanner at 0.25 um/px spacing, originally stored in MRXS file format. However, the WSIs made available for this project on Zenodo have been converted to TIFF format with a maximum spacing of 0.5 um/px. This was done to make slides broadly accessible (since MRXS files are sometimes not compatible with some digital pathology viewers or APIs) and with the same spacing used in the prediction of the pleomorphism score in [1].
The full list of files is the following:
AQ_S01_P000001_C0001.tif AQ_S01_P000002_C0001.tif AQ_S01_P000003_C0001.tif AQ_S01_P000005_C0001.tif AQ_S01_P000006_C0001.tif AQ_S01_P000007_C0001.tif AQ_S01_P000009_C0001_B217.tif AQ_S01_P000010_C0001.tif AQ_S01_P000011_C0001.tif AQ_S01_P000012_C0001.tif AQ_S01_P000013_C0001.tif AQ_S01_P000015_C0001_B208.tif AQ_S01_P000016_C0001.tif AQ_S01_P000018_C0001.tif AQ_S01_P000019_C0001.tif AQ_S01_P000020_C0001.tif AQ_S01_P000021_C0001_B106.tif AQ_S01_P000021_C0001_B116.tif AQ_S01_P000022_C0001.tif AQ_S01_P000024_C0001.tif AQ_S01_P000025_C0001.tif AQ_S01_P000027_C0001.tif AQ_S01_P000028_C0001.tif AQ_S01_P000029_C0001.tif AQ_S01_P000030_C0001_B102T1.tif AQ_S01_P000030_C0001_B102T2.tif AQ_S01_P000032_C0001_B203.tif AQ_S01_P000033_C0001.tif AQ_S01_P000035_C0001.tif AQ_S01_P000037_C0001.tif AQ_S01_P000038_C0001.tif AQ_S01_P000039_C0001.tif AQ_S01_P000045_C0001.tif AQ_S01_P000046_C0001.tif AQ_S01_P000048_C0001_B107.tif AQ_S01_P000048_C0001_B110.tif AQ_S01_P000049_C0001_B208.tif AQ_S01_P000050_C0001.tif AQ_S01_P000052_C0001.tif AQ_S01_P000053_C0001.tif AQ_S01_P000055_C0001.tif AQ_S01_P000056_C0001_B103.tif AQ_S01_P000056_C0001_B107.tif AQ_S01_P000056_C0001_B114.tif AQ_S01_P000058_C0001.tif AQ_S01_P000059_C0001.tif AQ_S01_P000062_C0001.tif AQ_S01_P000063_C0001_B103.tif AQ_S01_P000063_C0001_B105.tif AQ_S01_P000063_C0001_B302.tif AQ_S01_P000065_C0001_B103.tif AQ_S01_P000065_C0001_B106.tif AQ_S01_P000066_C0001.tif AQ_S01_P000067_C0001.tif AQ_S01_P000068_C0001.tif AQ_S01_P000069_C0001.tif AQ_S01_P000070_C0001.tif AQ_S01_P000071_C0001_B203.tif AQ_S01_P000072_C0001.tif AQ_S01_P000073_C0001.tif AQ_S01_P000074_C0001_B102.tif AQ_S01_P000074_C0001_B115.tif AQ_S01_P000075_C0001.tif AQ_S01_P000076_C0001.tif AQ_S01_P000077_C0001.tif AQ_S01_P000078_C0001.tif AQ_S01_P000079_C0001.tif AQ_S01_P000080_C0001.tif AQ_S01_P000083_C0001.tif AQ_S01_P000084_C0001_B105.tif AQ_S01_P000084_C0001_B111.tif AQ_S01_P000084_C0001_B214.tif AQ_S01_P000085_C0001.tif AQ_S01_P000086_C0001.tif AQ_S01_P000087_C0001.tif AQ_S01_P000089_C0001.tif AQ_S01_P000094_C0001.tif AQ_S01_P000095_C0001.tif AQ_S01_P000096_C0001.tif AQ_S01_P000098_C0001.tif AQ_S01_P000099_C0001.tif AQ_S01_P000100_C0001.tif AQ_S01_P000101_C0001.tif AQ_S01_P000102_C0001.tif AQ_S01_P000103_C0001.tif AQ_S01_P000104_C0001.tif AQ_S01_P000105_C0001.tif AQ_S01_P000106_C0001.tif AQ_S01_P000107_C0001.tif AQ_S01_P000108_C0001.tif AQ_S01_P000110_C0001.tif AQ_S01_P000111_C0001_B104.tif AQ_S01_P000112_C0001_B211.tif AQ_S01_P000115_C0001.tif AQ_S01_P000116_C0001.tif AQ_S01_P000120_C0001.tif AQ_S01_P000122_C0001_B212.tif AQ_S01_P000124_C0001_B218.tif AQ_S01_P000127_C0001.tif AQ_S01_P000133_C0001.tif AQ_S01_P000137_C0001.tif AQ_S01_P000141_C0001.tif AQ_S01_P000142_C0001.tif AQ_S01_P000147_C0001.tif AQ_S01_P000151_C0001.tif AQ_S01_P000175_C0001.tif AQ_S01_P000177_C0001.tif AQ_S01_P000178_C0001.tif AQ_S01_P000179_C0001.tif AQ_S01_P000183_C0001.tif AQ_S01_P000184_C0001.tif AQ_S01_P000185_C0001_B110.tif AQ_S01_P000185_C0001.tif AQ_S01_P000187_C0001.tif AQ_S01_P000189_C0001.tif AQ_S01_P000190_C0001.tif AQ_S01_P000191_C0001_B105.tif AQ_S01_P000191_C0001_B110.tif
References
[1] C. Mercan, M. Balkenhol, R. Salgado, M. Sherman, P. Vielh, W. Vreuls, A. Polonia, H. M. Horlings, W. Weichert, J. M. Carter, P. Bult, M. Christgen, C. Denkert, K. van de Vijver, J.-M Bokhorst, J. van der Laak, F. Ciompi, Deep learning for fully-automated nuclear pleomorphism scoring in breast cancer. Npj Breast Cancer, 2022.